Protein Barcodes and Next-Generation Protein Sequencing™
April 16, 2024 • Brian Reed, PhD

Game-changing technology for high-throughput, multiplexed assays
Brian Reed, Ph.D., Head of Research
Characterizing proteins and understanding changes to protein isoforms are essential components of biological research, proteomics studies, and the drug discovery workflow. Conventional techniques, however, are time- and labor-intensive, suffer from throughput constraints, and have other shortcomings. Protein detection assays such as western blots and ELISA, for example, are limited in their ability to resolve unknown mutations, truncations, and proteoforms. Although mass spectrometry can sometime resolve these differences, the technique requires specialized technical expertise and has a slow turnaround time.
Identifying protein variants is similarly challenging, as assays typically rely on large screens that prolong engineering cycles, restrict throughput, and extend the time to actionable data.
In the drug discovery setting, where it is especially critical to screen and characterize large numbers of proteins, time is of the essence. Low-throughput, laborious assays are problematic, slowing the overall workflow, and limiting how many proteins can be screened and characterized in a reasonable timeframe.
The combination of protein barcodes and a user-friendly platform to directly read and identify sequences with single-molecule resolution is a game-changing technology for researchers and drug discovery scientists as they screen and characterize proteins.
DNA Barcoding Set the Stage
DNA barcodes—short stretches of DNA used to encode information—have been applied in a wide array of applications thanks to the availability of next generation DNA sequencing (NGS). When used in combination with NGS, DNA barcoding allows tracking of sample identity in multiplexed libraries and single-cell resolution in transcriptomic studies, among many other applications. With four different nucleotides, a total of 410 (approximately 1 million) unique sequences can be generated, assuming a 10-letter barcode. NGS decodes the information contained with the DNA barcodes in a high-throughput, cost-effective manner.
Protein Barcoding Takes it to the Next Level
Protein barcodes are information-rich, short stretches of amino acids that can be genetically encoded into the coding sequences for proteins. This process yields a library of proteins with distinct barcodes, each of which can be accurately identified using next-generation protein sequencing (NGPS) on Quantum-Si’s Platinum® instrument.
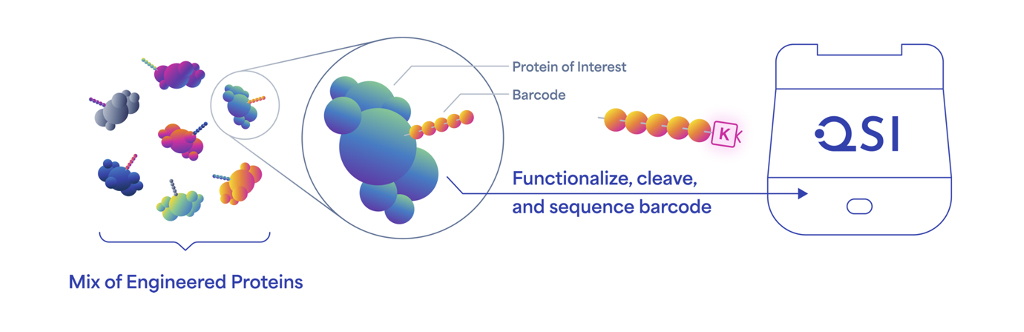
The Platinum® instrument is the first to enable protein barcode sequences to be read directly and identified with single-molecule resolution. Previously published studies using protein barcodes relied on mass spectrometry for decoding, but this technology can’t always differentiate among so many peptides of similar length and sequence.1 Mass spectrometry data are also complex and hard to interpret and analyze.
While protein barcodes operate in a similar manner to DNA barcodes, access to 20 amino acids, compared to four nucleotides, yields vastly more unique barcode sequences and a greater capacity to encode extensive information in short sequences. For example, a 10-letter protein barcode can encode 2010 (approximately 10 trillion) unique sequences.
See Proteins in a New Way
The combination of protein barcodes and Quantum-Si’s NGPS platform enables multiplexed protein variant screening and characterization studies, delivering significant increases in assay speed and throughput. The multiplex nature of these assays also reduces the number of cell culture plates or vessels that are needed, further streamlining workflows.
Here are just a few examples of how this technology can be applied.
Protein Trafficking
Many proteins reside in multiple subcellular regions with different proteoforms located in distinct areas and performing different biological roles.2 3 The movement of proteins through multiple organelles is important for signaling and regulating cellular physiology, and mis-localization can lead to dysfunction and diseases including cancer and neurodegenerative disorders.4
A library of barcoded protein variants engineered with mutations in the putative trafficking domain can be produced and expressed in a cell line. Following an incubation period, the various organelles, along with associated proteins with barcodes, can be isolated. Sequencing the barcodes will reveal the identity of the protein variants present in the respective organelles.
Protein – Protein Interactions
Protein-protein interactions play a critical role in many physiological and pathological processes. Aberrant interactions have been associated with many diseases, including cancer, infectious diseases, and neurodegenerative conditions,5 and modulation of these interactions has emerged as a target class for drug discovery.6
Protein barcoding provides important advantages when studying and pinpointing the specific locations at the protein-protein interface that dictate both normal and aberrant interaction. A large panel of variants containing mutations at the interface can be created, and each one tagged with a unique barcode. A multiplexed, high-throughput assay can then reveal how changes to a protein’s structure affect its ability to interact.
Protein Characterization
mRNA medicine, in which genetic information in the form of mRNA directs therapeutic protein production, is gaining traction as a novel drug modality.7 Development of these drug candidates typically involves generating a large number of mRNAs with iterations of the coding sequence, followed by screening to determine which sequences express the protein at the desired level.
By appending coding sequencing for protein barcodes to each mRNA coding sequence, expressed proteins will have unique tags, despite the proteins themselves having the same amino acid sequence. Sequencing the barcodes on the Platinum® NGPS platform can then allow rapid identification of which mRNA had the highest expression, resulting in the most abundant protein.
Ask and Answer New Questions
Combining protein barcodes with NGPS on the Platinum® instrument creates the opportunity for multiplexed protein characterization, variant screening, and many other applications with unprecedented speed and throughput. In addition to accelerating workflows, processing more samples in a single assay reduces costs and labor requirements.
The ability to directly read and identify protein barcodes with single-molecule resolution has the potential to transform research and drug discovery in a manner like that of DNA-based barcodes and NGS. This new tool will allow scientists to ask and answer new questions, setting the stage for new and unexpected discoveries, and a new understanding of health and disease. To learn more about protein barcoding, access our application note.

Brian Reed (Bio)
Brian Reed, Ph.D., is a distinguished scientist and the Head of Research at Quantum-Si, where he has been at the forefront of groundbreaking advancements. Dr. Reed is part of the team that developed Quantum-Si’s Platinum™ platform which includes the world’s first next-generation protein sequencing™ instrument.
Dr. Reed joined Quantum-Si in June 2014, assuming the role of Principal Scientist and was elevated to Head of Research in August 2019. Prior to Quantum-Si, Dr. Reed was a Senior Staff Scientist and Group Leader at Ion Torrent’s Life Technologies.
He earned his Bachelor of Arts in Biology at Harvard University and went on to earn his Ph.D. in Molecular Biology from Yale University.
References
- Egloff, P. et al. Engineered Peptide Barcodes for In-Depth Analyses of Binding Protein Ensembles. Nat.Methods 16, 421–428 (2019)
- Cook K.C., Cristea I.M. Location is everything: protein translocations as a viral infection strategy, Current Opinion in Chemical Biology. 48:34-43. 2019.
- Johannes L., Popoff V. Tracing the Retrograde Route in Protein Trafficking, Cell. 135(7): 1175-1187. 2008.
- Wang Y., Qin W. Revealing protein trafficking by proximity labeling-based proteomics. Bioorganic Chemistry, Volume 143:107041. 2024.
- Lu, H., et al. Recent advances in the development of protein–protein interactions modulators: mechanisms and clinical trials. Sig Transduct Target Ther 5, 213 (2020).
- Skwarczynska M., Ottmann C. Protein-protein interactions as drug targets. Future Med Chem. 2015;7(16):2195-219. doi: 10.4155/fmc.15.138. Epub 2015 Oct 29. PMID: 26510391.
- Metkar, M. et al. Tailor made: the art of therapeutic mRNA design. Nat Rev Drug Discov 23, 67–83 (2024).